1097-4547/asset/olalertbanner.jpg?v=1&s=ab2ec4e1133a4778a617826911b6060e49c6ffdf)
Sexism fears hamper brain research
Articles of interest to a medical doctor with interests in Psychiatry, Technology and Ophthalmology.













1748-7692/asset/mms_large.jpg?v=1&s=0089eb82c2de1dee2faffe4d2493372aa902082c)




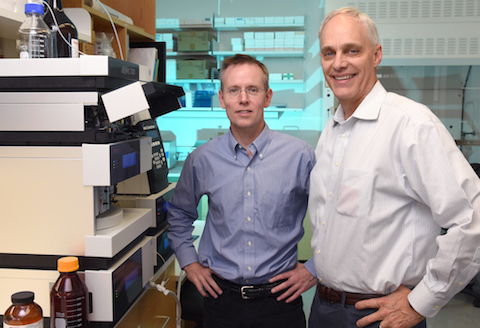
Hi reddit, my name is Daniel Irimia and I am an Associate Professor in the Surgery Department at Massachusetts General Hospital and Harvard Medical School, and a Senior Investigator at Shriners Burns Hospital in Boston. My research focuses on designing novel technologies for measuring the activities of white blood cells from patients, towards better ways to predict, diagnose, monitor, and treat inflammation, infections, and sepsis. In 2016, I received the "Pioneers of Miniaturization" prize from the Chemical and Biological Microsystems Society, for pioneering work on microfluidic technologies for measuring human neutrophil activities and applications to human diseases.
I am the organizer of the recent Dicty World Race, an unorthodox approach aimed at encouraging biologists to employ emerging microfluidic technologies to make high precision measurements of cell migration for biological and medical research applications. The results and learning from this experiment were recently published as an article titled “A Worldwide Competition to Compare the Speed and Chemotactic Accuracy of Neutrophil-Like Cells” in PLOS ONE. The race enabled a large-scale comparison of motility and chemotaxis in the engineered cell lines, allowing exploration of a diverse set of strategies for enhancing chemotactic performance. We found that there are tradeoffs between cell speed and chemotactic accuracy in maze-like environments and that the winning cells were not the fastest cell type, but excelled in finding the shortest paths through the maze. These findings could eventually help us develop better therapies against infections and chronic inflammation.
Don’t forget to follow me on Twitter [@D__Irimia](http://www.twitter.com/D__Irimia].
I will be answering your questions at 1pm ET -- Ask Me Anything!





















Hi Reddit!
New molecular engineering technology allows scientists to edit the DNA of a cell with greater accuracy than previous methods. This technology, called CRISPR-Cas9, has potential applications in agriculture and health.
To further improve the accuracy of CRISPR, we have developed GT-Scan2, a software tool that helps researchers identify the optimal CRISPR target site by crunching through large amounts of data and using epigenomic information to predict CRISPR activity. We implemented GT-Scan2 using a new Amazon Web Service technology only released this year, Lambda, which allows us to break down large tasks into smaller sub-tasks that can be solved in parallel on the cloud. GT-Scan2 was featured on the prestigious Jeff Barr blog because it brings together novel scientific insights and unprecedented cloud-compute capacity thereby democratizing high quality CRISPR target site search to power new research applications in the future.
Tell us what you think about the benefits of genome engineering and making target site search accessible to more researchers.
We will begin answering your questions at 5pm ET (9am Sydney), and we’re excited to hear from you. AUA!